
Searching for Biomarkers in Mass Spectra
When the software PATTERN EXPERT amspect is used in the field of
proteomics,
it is a valuable tool for searching biomarkers. Here, the term "biomarkers" refers to specific proteins, which can be used as
indicators for diagnosing diseases, identifying microorganisms, or doing other similar kinds of analysis. The knowledge of
good biomarkers for some disease, for example in cancer diagnozis, might even shed light on the underlying medical mechanisms.
Pattern recognition methods applied
to mass spectra can help to identify biomarkers. We will give a short description of this process:
Suppose, you wish to identify specific microorganisms and to find respective biomarkers. At first, you need to record a number of
mass spectra of samples of the given microorganisms as well as of a collection of other species, from which your target species is to be distinguished.
After that, the measured mass spectra are analyzed using
PATTERN EXPERT amspect.
A classification experiment is conducted trying to distinguish the different microorganisms. You may do a "blinded test" in order to obtain a realistic
estimate for the expected level of correctness when classifying unknown (i.e. untrained) spectra. If the blinded test yields a satisfactory result,
it is most interesting to analyze the set of features, which has been used to achieve it.
The program delivers a list of those spectral intervals, which have been used for classification. Now, these intervals can be associated with
biomarkers for the given classification task.
The software uses a series of powerful algorithms for selecting useful and relevant biomarkers.
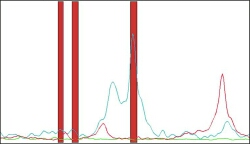 The diagram shows three spectral intervals (biomarkers) selected by
The diagram shows three spectral intervals (biomarkers) selected by
PATTERN EXPERT amspect.

